Every year, millions of Canadians experience the onslaught of a well-known and common enemy, the flu. The flu is one of many viruses that people can catch during flu season, and one of the viruses responsible for disrupted plans and miserable days. But humans are not the only creatures susceptible to viral infection – even plants can get ‘under the weather.’
From a virus’s perspective, plants and mammals have the same underlying machinery that can allow a virus to reproduce. Both have cells that can be hijacked and molecular tools that can be used for the hijacking; however, plant viruses have specifically adapted to the cellular biology of various plant species. This means that plant viruses are not capable of directly causing sickness in humans; however, that does not mean they do not significantly and negatively affect human health and livelihood.
Plant viruses can pose a significant threat to both crop yield and quality as they interfere with normal plant growth and development. They are often transmitted from crop to crop by insects, known as insect-vectored viral pathogens, which can spread throughout the crop in growing regions worldwide. Notably, some of the areas most susceptible to crop virus infection are located in North America, specifically in North Dakota and the Pacific Northwest. These regions have a long and contentious history of several plant viruses, including Barley Stripe Mosaic Virus (BSMV) and Soybean Mosaic Virus (SMV). In the 1950s, BSMV was widespread in virtually all barley fields in North Dakota, resulting in a 25% reduction of crop yield in affected fields. While the implementation of a strong seed testing program reduced the levels of BSMV in North Dakota to virtually zero in 1957, virus infection has remained a concern in these regions and others.
Present-day plant viruses continue to infect pea, lentils, faba, chickpeas, soybean, and occasionally alfalfa. In Saskatchewan, plant viruses have not generally been considered a significant threat to crop production, and currently, there is no estimate of the potential loss from virus infection in Western Canada. This historical lack of concern could potentially stem from minimal losses of pulse crops due to virus infection, or it could also be attributed to a lack of adequate detection methods. However, given that viruses, including Pea seed-borne mosaic virus (PSbMV), are routinely detected in North Dakota and Montana, which border Saskatchewan, and in the Palouse region, which borders Alberta, this is likely to change.
Therefore, Saskatchewan Pulse Growers (SPG) supported research, principally conducted by Dr. Sean Prager, that was initiated to generate tools and information necessary if or when viral infection becomes prevalent in Western Canada. This research had two goals, (1) to obtain information relating to the prevalence of major pulse crop viruses in Saskatchewan and (2) to develop Polymerase Chain Reaction (PCR) based detection techniques for these viruses, which include PSbMV, Pea streak virus (PeSV), Bean yellow mosaic virus (BYMV), Bean leafroll virus (BLRV), Alfalfa mosaic virus (AMV), and Cucumber mosaic virus (CMV).
This project employed an integrated approach to improve the detection, surveillance, and understanding of virus infections in Saskatchewan pulse crops. To develop virus detection tools, primers and probes were designed that could bind to the genetic code of plant viruses, allowing said code to be used for virus identification. These tools were validated through molecular screening of plant tissue, seeds, and insect vectors to identify known and novel viruses in symptomatic samples of pea, chickpea, lentil, and faba bean.
To assess virus prevalence and vector involvement, field surveys were conducted in 2023 and 2024 across major pulse-growing regions in Saskatchewan. Plant samples (both symptomatic and asymptomatic) and aphid populations were collected. PCR and Next Generation Sequencing (NGS) were then used to detect a panel of viruses from these samples. In controlled greenhouse settings, the mechanical inoculation of PSbMV and BYMV into several crop varieties enabled the documentation of symptom development, plant growth metrics, and yield impact. Pea aphid vector transmission assays were conducted to evaluate the efficiency of virus spread under varying aphid densities. Finally, genetic analysis of PSbMV was performed using conventional PCR and genome sequencing. Isolates from multiple years and regions were compared phylogenetically to global strains to understand the diversity and evolution of PSbMV across North America. Collectively, these methods provided a framework for virus detection, epidemiological surveillance, and risk assessment in pulse crop systems.
At the beginning of this project, information regarding the presence of pulse-infecting viruses in Saskatchewan was minimal. By the end of the project, the presence of multiple viruses was confirmed using the developed materials, including BLRV in peas and chickpeas, BYMV in beans and peas, endornaviruses in beans, PESV in pea and chickpeas, and PSbMV in faba beans and chickpeas. The locations of these positive identifications ranged throughout Southern Saskatchewan, with positives of BYMV also being detected in Bow Island, Alberta. Amongst these viruses, PSbMV, BLRV, BYMV, and endornaviruses have the potential to cause significant yield loss. However, endornaviruses are exclusively transmitted from parent to offspring; therefore, there are no reports of disease caused by these viruses in this region so far.
Next-generation sequencing was then used to estimate the incidence rates of the viruses within purified samples collected, and these results were compared to field symptom survey results. Based on field symptoms, viral incidence rate was predicted to be greater than 5% for BLRV, BYMV, and endornaviruses, and less than 5% for PeSV. Sanger sequencing enabled the confirmation of positive viral samples with greater certainty and allowed for comparison with available viral sequences to identify similarities. This process resulted in the first identification of BLRV from the pulse-growing region of the Canadian prairies, originating from two different hosts located in two unrelated fields, which suggests the possible circulation of this virus among pulse crops. From these results, more targeted trials were conducted. Specifically, disease symptoms observed in the field were reproduced in healthy bean plants by inoculating them with sap from infected field plants. Symptoms in the healthy plants appeared ten days after inoculation, and included yellowing, deformation, and necrosis. Using next-generation sequencing, BYMV was identified.
This project also aimed to identify the genetic diversity of PSbMV. Multiple isolates of PSbMV were isolated and sequenced. Comparing the sequences revealed an overall low genetic diversity of PSbMV in Saskatchewan, suggesting that there has been limited introduction of new strains in the province. The strain identified in Saskatchewan was determined to be closely related to those from North Dakota and Washington, implying a common origin of the viruses in these diverse regions. Overall, the above results demonstrated that several viruses of relevance to pulse crops are present in Saskatchewan, and that some of these viruses, such as Bean Yellow Mosaic Virus (BYMV), are infectious to healthy plants. Therefore, the research aimed to quantify further the yield effects of BYMV and PSbMV in important pulse crop varieties, as well as document the symptoms of these viruses on Saskatchewan-grown pulse crops.
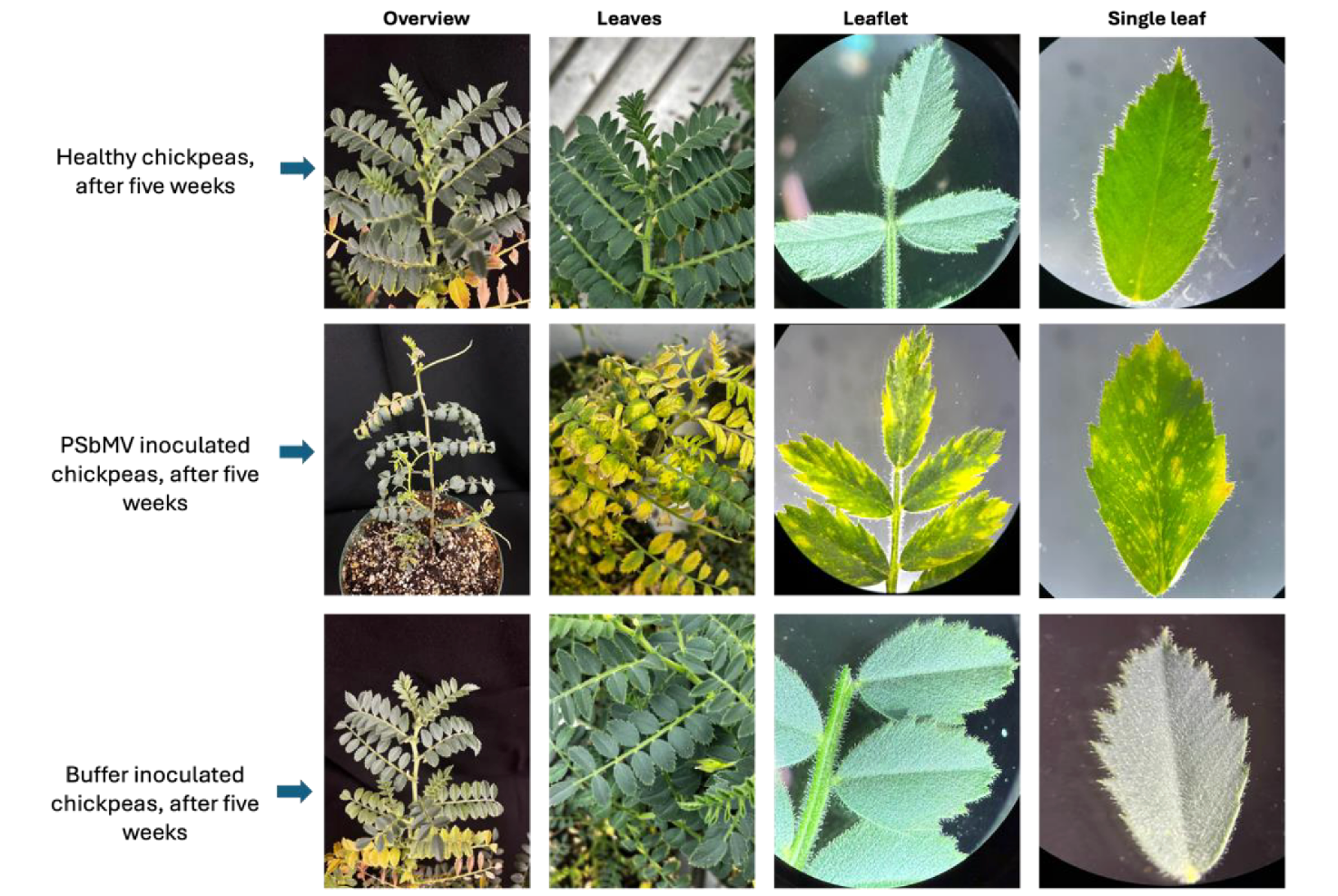
PSbMV was shown to cause significant and crop-specific effects in chickpea, faba bean, and pea. In chickpea, infection resulted in mosaic leaf patterns, yellowing, leaflet bunching, and tip necrosis, with infected plants failing to produce any pods, indicating complete reproductive failure (Figure 1). These plants also exhibited a nearly 50% reduction in height compared to buffer-treated and healthy controls. In contrast, PSbMV-infected faba bean plants showed symptoms such as excessive venation, yellowing, and mosaic leaf patterns (Figure 2). Although pod formation remained unaffected, the seeds were often smaller, misshapen, wrinkled, and mottled, suggesting that PSbMV primarily affects seed quality. While PSbMV was detected in the seed coat of infected faba bean seeds, no seed-to-seedling transmission occurred in germination trials. The virus was also consistently detected in single aphids and aphids stored under various conditions for up to 40 days, demonstrating its environmental persistence and vector association.

In pea, PSbMV infection manifested with a progression of symptoms, beginning with mild necrosis at leaf tips within 7 to 14 days post-inoculation (Figure 3). Symptoms intensified by 14–25 days, including pronounced leaf crinkling, curled leaf edges, severe mottling, chlorosis, burned tendrils, and deformities in flowers and pods. Pod formation declined, and seeds were frequently shrivelled or malformed. Under stressful conditions, infection led to widespread yellowing, necrosis, and complete dieback of the plant. Similarly, detailed symptom progression was recorded for BYMV in inoculated pea plants. At 7 days post-inoculation, mild chlorotic spots and vein clearing appeared on young leaves, with minimal growth impact. By 14 days, distinct mosaic patterns and yellowing emerged, along with early growth reduction. At 18 days, leaves showed crinkling, distortion, and rolling. By day 25, leaf deformation and excessive curling were severe, and overall plant growth was noticeably stunted. At 28 days, necrotic lesions and leaf tissue death began to appear, with wrinkled and brittle foliage. By 31 days post-inoculation, infected plants reached a terminal stage, exhibiting widespread necrosis, desiccation of leaves, tendrils, and stems, and complete plant collapse.

Finally, to evaluate the vector capacity of pea aphids in spreading viruses, transmission assays were conducted using BYMV- and PSbMV-infected aphids on chickpea and faba bean. Results showed that even a single infected aphid was sufficient to transmit the virus to both crops, demonstrating the high efficiency of nonpersistent virus transmission. In faba bean, infection rates reached nearly 100% when 10 or more aphids were present, indicating that viral spread correlates with aphid density. In contrast, chickpea exhibited lower transmission efficiency, with only 50% of plants exposed to five aphids testing positive. These findings confirm that pea aphids are highly effective vectors of BYMV and PSbMV, highlighting the need for vector management as a critical component of virus control strategies in pulse crops.
Overall, field surveys conducted from 2023 to 2024 confirmed the presence of several pulse crop viruses in Saskatchewan, notably detecting Bean Leaf Roll Virus (BLRV) in peas and chickpeas for the first time, underscoring the importance of continued monitoring. PSbMV was shown to transmit efficiently through seed in faba beans, with 100% of seeds from infected plants testing positive, highlighting the critical need for seed health screening protocols. Characteristic PSbMV symptoms were documented across crops, including burning tendrils and leaf curling in peas, and mosaic patterns and yellowing in faba beans, though symptom expression was suppressed under cooler temperatures.
The results suggest that strengthening virus surveillance programs, particularly in regions with known virus prevalence, and implementing regular aphid population monitoring can mitigate the spread of these viruses. Given the high rate of PSbMV seed transmission, seed lots should also be rigorously screened, and efforts should be made to develop or adopt certified virus-free seeds. Integrated pest management strategies targeting aphid vectors, through the use of biological controls and selective pesticide applications, can also be prioritized. Finally, early-season diagnostic testing is essential even when plants appear asymptomatic. There remains a need to determine the full viral diversity within Saskatchewan and to identify further the presence of potentially novel or emerging viruses that are currently undetectable using modern techniques. Additionally, there is very little known about the details surrounding virus distribution and temporal dynamics of infection, the host range of these viruses, and any mitigating host factors that lead to plant resistance. Lastly, and potentially most importantly, there is still a need for a comprehensive assessment of the virus-associated yield loss and economic damage to pulse crops.